As part of the TrainMALTA EU project activities, I volunteered/was tasked with setting up the IT infrastructure for the HTS (or NGS) bioinformatics summer school. It has been quite an experience, and the whole setup is far from trivial – so I thought I’d document parts of it here. Habitually, I turned to google to search what others in my shoes have done and nothing turned up. Nothing on google – this setup must be worth documenting!
We required a rig which fulfils the following criteria:
- Supports ~40 participants
- Supports six tutors, with their diverse software (and genomic storage) requirements
- Easy to propagate changes/software across to everyone
- Cheap(ish) – doh!
- Requires minimal participant setup (ideally none). We want to run the whole programme via a web browser.
- Is tried and tested in such a classroom environment- so as not to get any surprises on the day (when it’s too late…)
- Have a backup system in case of hiccups
Luckily our project partners at the University of Cambridge and Umeå (Sweden) have ample experience running these sort of courses (thanks Bastian!, all of the below is his setup). They immediately suggested we go for a cloud solution (Amazon Web Services), which suited us fine as local resources for this kind of course are limited, at best. The architecture of the system they suggested is shown pictorially below.
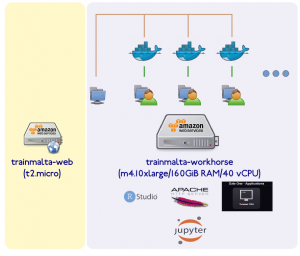
There are two main machines – a web host or gateway which allows participants to easily access bioinformatics software and materials, and a workhorse which does all the heavy lifting.
The webhost is a t2.micro (free) AWS instance which runs a node.js web server. A web page using Jade templating engine give a list of participants with links to the hosted services (shown below).
The workhorse machine is a m4.10xlarge AWS instance ($2.64/hr) with 160 GiB RAM and 40 vCPUs – ample for our 35+ students. Only the tutors have physical access to this machine (best to create individual user accounts and a tutors group too). The idea here is that we run a docker container for every participant, with services running on different ports for each user (details later).
On the workhorse we create a data/ directory (in the root folder, /) and subfolders for each day of the course (e.g. day1/, day2/, day3/, day4/, day5/). A further subdivision for each day could be slides/ and practicals/, where the former contains the course materials and the latter contains the data used for hands on practicals and exercises. This data directory is mounted directly (and in read-only mode) in each container instance, one per user using docker’s -v mounting option. On the workhorse we also mount a 5000 GiB amazon volume under /participants. Here we create as many home directories as we have participants (01/, 02/, 03/, etc.).
We start off with a docker image (Bastian’s) which is conveniently publicly available, and which you can retrieve with docker pull bschiffthaler/ngs. This image is used to start the docker container. We start by running a container, with port forwarding for ssh, http (an apache instance is running in the image), and RStudio. The container is started using port suffix 00, in the following manner:
docker run -d -p 10000:80 -p 10100:443 -p 2200:22 -p 8800:8888 -v /data:/data -v /participants/00:/home/training 7f5a79c3410d
Port 10000 of the container now runs apache, 10100 runs Gate One (a browser-based ssh client), 10200 runs RStudio (training/training login), and port 2200 runs an ssh service to the container. In the command above note the mounting of /data (from workhorse to container) and training user’s container home directory from /participants/00. On the workhorse, create a user participant_docker (with uid 2000 — this is a requirement) and use this as the home directory owner. Note that user training’s uid is also 2000, so this enables the training user to write in his/her home directory.
As always, training requirements differ between courses so the tutors will almost certainly need to each add and install their own bits of kit in the container. One way to do this is to let each of the tutors login (as root !) in the container (using something like docker exec -it dcd97a21424e /bin/bash). This allows them to apt-get install or pip install any other software they would like. Note that all tutors should be in group docker (to connect to the container). After finalizing the software installs you need to save (commit) to the image – this is simple enough to achieve (e.g. docker commit dcd97a21424e trainmalta2016_v4). A more maintainable way to do this is to edit the docker file and make all software readily available in the image, but if you are in a hurry… When the tutors finish their software+data installs we copy the training user home directory to each of participant directories in 01/, 02/, 03/, etc.
You can launch a container for each participant using a bash script like:
#!/bin/bash
IMAGEID=7f5a79c3410d
PARTICIPANTS=50
for f in $(eval echo "{1.."$PARTICIPANTS"}")
do
if [ $f -lt 10 ]
then
f="0$f"
fi
echo "Creating container for participant $f ... "
docker run -d -p 100$f:80 -p 101$f:443 -p 102$f:8787 -p 22$f:22 -p 88$f:8888 -v /data:/data -v /participants/$f:/home/training $IMAGEID /usr/bin/supervisord -c /etc/supervisor/conf.d/supervisord.conf
done
The supervisor daemon (supervisord) needs to be run because the list of startup programs has changed from the original image to include jupyter notebook.
This worked well for most of our training needs with a few exceptions. For example, we wanted to show the participants the IGV browser. However this launches its own GUI (workhorse runs in headless mode). Participants were asked to install IGV locally (on their laptops or on the classroom’s permanent machines) and then just download the file of interest from the apache directory listing (via the browser). More advanced participants used sshfs to mount the directory locally from the workhorse. A workaround could be to launch ssh to the workhorse with X tunnelling (perhaps?).
The summer school is mid-way through and (hopefully without jinxing it) has gone well so far.
Update, July 2018
We used pretty much a similar setting for the TrainMALTA Summer School in epigenetics (2018). I noticed that there were some important points missing, mainly:
- modules should be installed in
docker_training‘s home directory which will be mounted in the docker instance (i.e.traininguser home dir) docker_traininguser in the workhorse has the same uid (2000) as thetraininguser in the docker instance- The
docker_traininghome directory contents should be copied in each user’s dir (e.g./participants/00,/participants/01, etc.) - Code for the front end is downloaded from – https://microasp.upsc.se/bastian/course-web
- The front-end is a node server with jade templating, you will need to edit pages for materials, programme, student lists, connect to AWS instance, etc
- To run the front end server you need to
run DEBUG="untitled1:server" nodejs ./bin/www
Leave a Reply